Preliminaries
Introduction
So, to begin, what is R and why should we use R for spatial analysis? Let’s break that into two questions - first, what is R and why should we use it?
- A language and environment for statistical computing and graphics
- R is lightweight, free, open-source and cross-platform
- Works with contributed packages - currently 12,938 -extensibility
- Automation and recording of workflow (reproducibility)
- Optimized work flow - data manipulation, analysis and visualization all in one place
- R does not alter underlying data - manipulation and visualization in memory
- R is great for repetetive graphics
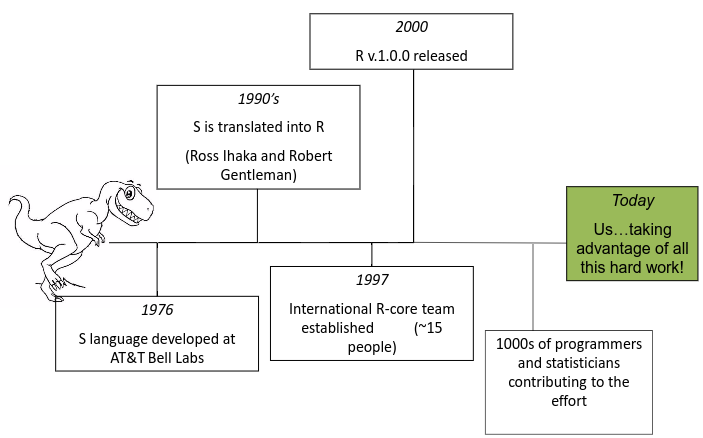
History of R
Second, why use R for spatial, or GIS, workflows?
- Spatial and statistical analysis in one environment
- Leverage statistical power of R (i.e. modeling spatial data, data visualization, statistical exploration)
- Can handle vector and raster data, as well as work with spatial databases and pretty much any data format spatial data comes in
- R’s GIS capabilities growing rapidly right now - new packages added monthly - currently about 180 spatial packages
Some drawbacks to using R for GIS work
- R not as good for interactive use as desktop GIS applications like ArcGIS or QGIS (i.e. editing features, panning, zooming, and analysis on selected subsets of features)
- Explicit coordinate system handling by the user, no on-the-fly projection support
- In memory analysis does not scale well with large GIS vector and tabular data
- Steep learning curve
- Up to you to find packages to do what you need - help not always great
An ideal solution for many tasks is using R in conjunction with traditional GIS software.
R runs on contributed packages - it has core functionality, but all the spatial work we would do in R is contained in user-contributed packages. Primary ones you’ll want to familiarize yourself with are sf, rgdal, sp, rgeos, raster - there are many, many more. A good source to learn about available R spatial packages is:
Necessary R packages
First we need to install several R packages. Note the use of the terms package and library in R - you encounter both, and if you want to delve into semantics of which to use see this post on R-bloggers. R operates on user-contributed packages, and we’ll be jumping into use of several of these spatial packages in this workshop. Several packages we’ll be making use of are sp, rgdal, rgeos, raster, and the new sf simple features package by Edzer Pebesma. You should be able to use the packages tab in RStudio (see below) to install packages in a straightforward way. Mac and Linux users may have certain pre-requisites to fill, I’ll assume you can navigate these on your own or can assist as needed.
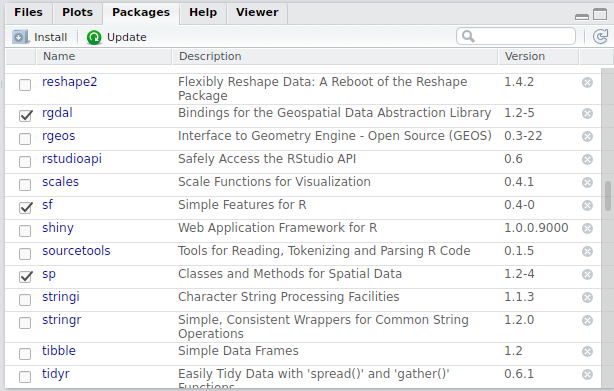
RStudio Console
Install all of the following packages in R: note that for both UPDATE: You can simply use the current CRAN release of sf and tidyverse - and specificallly ggplot2 in tidyverse, I’ve indicated the alternative install from GitHub rather than CRAN. This is optional, as is installing devtools, and you will be fine with the CRAN version of packages, except that you will not be able to reproduce one of the example plots in the sf section that uses sf_geom funtion from the development version of ggplot2.ggplot2 without using the devtools install of github - you’ll just want to ensure you are using ggplot2 >= 3.0.0 by running library(ggplot2) and sessionInfo() at your R console - within info returned you should see ‘ggplot2_3.0.0’ or higher. Note that tidyverse is a ‘meta-package’ that includes several specific packages such as ggplot2, dplyr, and tidyr.
install.packages("devtools") # optional but needed for using install_github
install.packages("rgdal")
install.packages("rgeos")
install.packages("raster")
# From CRAN:
install.packages("sf")
# From GitHub:
# library(devtools)
# devtools::install_github("r-spatial/sf")
# if you are running 3.5.1 on windows and have trouble with devtools install, try:
# assignInNamespace("version_info", c(devtools:::version_info, list("3.5" = list(version_min = "3.3.0", version_max = "99.99.99", path = "bin"))), "devtools")
install.packages("maptools")
install.packages("stringr")
install.packages("reshape")
install.packages("tidyverse")
install.packages("micromap")
install.packages("tmap")
install.packages("RCurl")
install.packages("dataRetrieval")
install.packages("maps")
install.packages("USAboundaries")
install.packages("rasterVis")
install.packages("landsat")
# From GitHub
# install.github("ropensci/plotly")
install.packages("plotly")
install.packages("leaflet")
install.packages("lubridate")
install.packages("tidycensus")
install.packages("rnaturalearth")
install.packages("osmdata")
install.packages('FedData')
install.packages("mapview")
# From GitHub
# devtools::install_github("r-spatial/mapview@develop")
install.packages("cranlogs")If EPA folks have any trouble with installing the CRAN version of mapview with the agency current version of R you can try using devtools to install the prior CRAN binary package of mapview described here.
Installing rgdal will install the foundation spatial package, sp, as a dependency, and installing tidyverse will install both ggplot2 and dplyr.
For Linux users, to install simple features for R (sf), you need GDAL >= 2.0.0, GEOS >= 3.3.0, and Proj.4 >= 4.8.0. Edzer Pebesma’s Simple Features for R GitHub repo has a good explanation:
You basically want to add ubuntugis-unstable to the package repositories and then get those three dependencies:
sudo add-apt-repository ppa:ubuntugis/ubuntugis-unstable
sudo apt-get install libgdal-dev libgeos-dev libproj-devThe Simple features for R package , sf, also needs udunits and udunits2 which may need coercing in linux:
Units Issues in sf GitHub repo
The following should resolve:
Basic R Background
Terminology: Working Directory
Working directory in R is the location on your computer R is working from. To determine your working directory, in console type:
## [1] "C:/Users/mweber/GitProjects/R-User-Group-Spatial-Workshop-2018"Which should return something like:
To see what is in the directory:
## [1] "_site.yml"
## [2] "data"
## [3] "GADM_2.8_USA_adm2.rds"
## [4] "header.html"
## [5] "img"
## [6] "index.html"
## [7] "index.Rmd"
## [8] "Mapping.html"
## [9] "Mapping.Rmd"
## [10] "Mapping_files"
## [11] "Preliminaries.html"
## [12] "Preliminaries.Rmd"
## [13] "R-User-Group-Spatial-Workshop-2018.Rproj"
## [14] "RAW"
## [15] "README.html"
## [16] "README.md"
## [17] "readme.txt"
## [18] "Resources.html"
## [19] "Resources.Rmd"
## [20] "site_libs"
## [21] "SpatialObjects.html"
## [22] "SpatialObjects.Rmd"
## [23] "SpatialObjects_files"
## [24] "SpatialOperations1.html"
## [25] "SpatialOperations1.Rmd"
## [26] "SpatialOperations1_files"
## [27] "SpatialOperations2.html"
## [28] "SpatialOperations2.Rmd"
## [29] "SpatialOperations2_files"
## [30] "srtm_12_04.hdr"
## [31] "srtm_12_04.tfw"
## [32] "srtm_12_04.tif"
## [33] "state_county_boundary.gdb"
## [34] "state_county_boundary.zip"
## [35] "style.css"To establish a different directory:
Terminology: data structures
R is an interpreted language (access through a command-line interpreter) with a number of data structures (vectors, matrices, arrays, data frames, lists) and extensible objects (regression models, time-series, geospatial coordinates) and supports procedural programming with functions.
To learn about objects, become friends with the built-in class and str functions. Let’s explore the built-in iris data set to start:
## [1] "data.frame"## 'data.frame': 150 obs. of 5 variables:
## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...As we can see, iris is a data frame and is used extensively for beginning tutorials on learning R. Data frames consist of rows of observations on columns of values for variables of interest - they are one of the fundamental and most important data structures in R.
But as we see in the result of str(iris) above, following the information that iris is a data frame with 150 observations of 5 variables, we get information on each of the variables, in this case that 4 are numeric and one is a factor with three levels.
First off, R has several main data types:
- logical
- integer
- double
- complex
- character
- raw
- list
- NULL
- closure (function)
- special
- builtin (basic functions and operators)
- environment
- S4 (some S4 objects)
- others you won’t run into at user level
We can ask what data type something is using typeof:
We see a couple interesting things here - iris, which we just said is a data frame, is a data type of list. Sepal.Length is data type double, and in str(iris) we saw it was numeric - that makes sense - but we see that Species is data type integer, and in str(iris) we were told this variable was a factor with three levels. What’s going on here?
First off, class refers to the abstract type of an object in R, whereas typeof or mode refer to how an object is stored in memory. So iris is an object of class data.frame, but it is stored in memory as a list (i.e. each column is an item in a list). Note that this allows data frames to have columns of different classes, whereas a matrix needs to be all of the same mode.
For our Species column, We see it’s mode is numeric, it’s typeof is integer, and it’s class is factor. Nominal variables in R are treated as a vector of integers 1:k, where k is the number of unique values of that nominal variable and a mapping of the character strings to these integer values.
This allows us to quickly see see all the unique values of a particular nominal variable or quickly re-asign a level of a nominal variable to a new value - remember, everything in R is in memory, so don’t worry about tweaking the data!
See if you can explain how that re-asignment we just did worked.
To access particular columns in a data frame, as we saw above, we use the $ operator - we can see the value for Species for each observation in `iris by doing:
To access particular columns or rows of a data frame, we use indexing:
A handy function is names, which you can use to get or to set data frame variable names:
Explain what this last line did
Overview of Classes and Methods
- Class: object types
class(): gives the class typetypeof(): information on how the object is storedstr(): how the object is structured
- Method: generic functions
print()plot()- `summary()
Workshop Data and Logistics
All the material for this workshop is in a GitHub repository.
There are two simple ways to get all the material for the course on your local machine:
- If you are comfortable with git and GitHub, you should clone the repository locally, and then you’ll have everything
- If git and GitHub are new to you, on the workshop repository page, click the green ‘Clone or download’ button, and choose ‘Download ZIP’. This will download everything in the workshop repository as a zip file for you locally.
For the workshop, the way things will run is:
- We will follow along with the content on the workshop web pages which I will have up on the screen (and you can have up on your machine as well)
- We will all have RStudio with R installed and running
- Each of the workshop sections is an RMarkdown file - if you have never used
RMarkdown, take a few minutes to explore RMarkdown or look over the nice overview put together by Ryan Hill and Marcus Beck for their recent R Spatial SFS Workshop. - Code in an
RMarkdowndocument will run just the same as code in an R script. I do all my work in .Rmd files rather than .r files in order to easily share work, create attractive output and reports weaving together code, images, figures and documentation, and follow a reproducible workflow. - As we go through each section, you should have the corresponding .Rmd file open for each section so that you can run and replicate the code, and have a another temporary R file open to solve questions, try your own approaches, and explore ideas presented.
- You are welcome to move through content at your own pace if you are already comfortable with material, but be aware that much of the code needs to be run in a sequential fashion in order to run (so that you have correct objects in memory)
- The course pages use ‘code-folding’ - at the top of the page you have the option to show all code, but ideally you will unfold code to run each section and in many sections try on your own prior to undolfing the code chunk to see the solution.
Dr. Wei-Lun Tsai has graciously offered to assist in the workshop, and will help with quesitons that arise, and Dr. Michael McManus will be helping remote participants with any quesitons and monitoring the chat in Skype. If we run into technical difficulties with remote participants, Mike McManus can be reached at his office line (513-569-7994) to help individuals with connection problems, and if we have major problems with Skype, we can use this call-information: 866.299.3188 Conf. Code 541.754.4469.